This set of Bioseparation Processes Multiple Choice Questions & Answers (MCQs) focuses on “Organic Solvent Precipitation”.
1. What is the governing equation for solvent based precipitation of protein?
a) ln\((\frac{S}{S_w}) = (\frac{A}{RT}) ((\frac{1}{e_w}) – (\frac{1}{e}))\)
b) ln\((\frac{S}{S_w}) = (\frac{A}{RT}) ((\frac{1}{e_w}) + (\frac{1}{e}))\)
c) ln S = β’ – K’ cp
d) ln S = β – K’ I
View Answer
Explanation: The governing equation for solvent based precipitation of protein ln \((\frac{S}{S_w}) = (\frac{A}{RT}) ((\frac{1}{e_w}) – (\frac{1}{e}))\). The organic solvents precipitate proteins and other molecules by the reducing the dielectric constant of the solution of the fermentation broth. In this equation, S is the solubility of the protein, Sw is the solubility of protein in water, A is the constant, e is the dielectric constant of the medium and ew is the dielectric constant of water.
2. Can higher concentration of organic solvents denature protein?
a) False
b) True
View Answer
Explanation: The higher concentration of organic solvents can denature the proteins and the organic solvents binds at the specific locations on the protein molecule and thus disrupts the hydrophobic interactions which hold the protein molecules in place. Therefore small amount of organic solvents are used in precipitation processes and these are carried out at low temperatures to minimize denaturation.
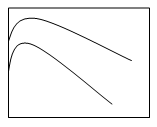
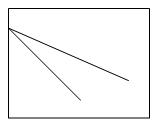
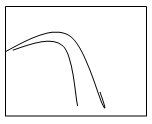
3. The solubility-concentration plot for protein precipitation using anti-chaotropic salt is ___
a)
b)

c)
d)
View Answer
Explanation: The protein precipitation plot expressing solubility and concentration of anti-chaotropic salts like ammonium sulphate is shown by graph ‘a’ which exposes hydrophobic patches on proteins by removing the highly structured water layer which covers the patches in the fermentation broth.
4. Is initial mixing required to achieve homogeneity for precipitation?
a) False
b) True
View Answer
Explanation: Initial mixing is important to achieve the homogeneity for precipitation; it can possible by adding a component like salts or any solvent which can initiate the process of precipitation. It is important to bring precipitant and the product molecules into collision.
5. What is Kolmogoroff model?
a) Macro-mixing
b) Mixing
c) Micro-mixing
d) Addition of solvents
View Answer
Explanation: Kolmogoroff model was studied in the context of micro-mixing for precipitation, in the form of homogeneous isotropic turbulence model. It assumes that mixing the product and the solutes in the solution are randomly dispersed eddies and these mixing is limited by diffusion.
6. What is Kolmogoroff length?
a) Length of different eddies
b) Differential length of eddies
c) Integral length of eddies
d) Mean length of eddies
View Answer
Explanation: Kolmogoroff length is the mean length of eddies and is designated by le. le = (\(\frac{\rho v^3}{P⁄V})^{1/4}\) where, ρ is the density of liquid, v is the kinematic velocity of liquid, P⁄V is the agitator power input per unit volume of liquid.
7. How to estimate the time required for mixing all the molecules to diffuse across all eddies?
a) t = \(\frac{\delta^2}{2D}\)
b) t = \(\frac{l_e^2}{8D}\)
c) t = –\(\frac{\delta^2}{2D}\)
d) t = –\(\frac{l_e^2}{8D}\)
View Answer
Explanation: The estimation of time required for the mixing all the molecules to diffuse across all eddies is performed by t = \(\frac{\delta^2}{2D}\) where, δ is the distance covered by diffusion of the molecules and D is the diffusion coefficient for the molecules being mixed. For spherical eddies t = \(\frac{l_e^2}{8D}\) is used where, le is the diameter for spherical eddies.
8. Estimate the concentration of nuclei at the end of initial mixing when 200l of aq. Solution contains 0.5g/l of macroglobulin at 20°C and 6 × 10-4 cm2/sec diffusion coefficient and the density of precipitate is 1.3g/cm3. Given: 95W is the power of stirring tank and mol. Wt. of macroglobulin is 9000000, t is 10s, Lmol = 5 × 10-6 cm.
a) 1.0 × 1010 \(\frac{particles}{cm^3}\)
b) 1.33 × 109 \(\frac{particles}{cm^3}\)
c) 1.54 × 1020 \(\frac{particles}{cm^3}\)
d) 1.0 × 1020 \(\frac{particles}{cm^3}\)
View Answer
Explanation: The power per unit volumes \(\frac{P}{V} = (\frac{95W}{200l})(\frac{10^7 g cm^2 s^{-3}}{W})(\frac{1l}{1000 cm^3}\)) = 47.5 × 103 \(\frac{g cm^2 s^{-3}}{cm^3}\) and the time required for the particles to reach eddies is 10 s. Since diffusion is limiting during this 6 s period, the number of concentration of nuclei N can be determined from the integrated form of second-order rate equation for the growth of particles limited by diffusion N = \(\frac{1}{K_A^t + 1/N_0}\) where, KA = 8πDLmol and Lmol = 5 × 10-6 cm so, KA = 8π\((\frac{6 × 10^{-7} cm^2}{s})\) (5 × 10-6 cm) = 7.54 × 10-11 cm3/s. N0 = \(\frac{0.5g}{l(\frac{l}{1000 cm^3})(\frac{mol}{820,000g})(6.02 × \frac{10^{23} molecules}{mol})}\) = 3.67 × 1014 cm-3 ∴ N = [(7.54 × 10-11 \(\frac{cm^3}{s}\))(10s) + (3.67 × 1014 cm-3)-1]-1 = 1.33 × 109 particles/cm3.
9. What will be the concentration of the protein when the particle is 1.0μm and the time for the particle to reach a size of 1.0μm?
a) 5.85min
b) 6.45min
c) 5.24 min
d) 6.75min
View Answer
Explanation: The ratio of the molecular weight of the particle to that of an individual molecule, \(\frac{M}{M_0}\) will be used to calculate the time to reach this particle t = \(\frac{\frac{M}{M_0} – 1}{K_A N_0}\), \(\frac{volume}{particle} = \frac{\pi(10^{-4} cm)^3}{6}\) = 5.23 × 10-13 cm3, \(\frac{mass}{particle}\) = (5.23 × 10-13 cm3)(\(\frac{1.3g}{cm^3}\)) = 6.80 × 10-13 g, \(\frac{molecules}{particle}\) = (6.80 × 10-13g)((\(\frac{mol}{820,000g})\)(6.02 × \(\frac{10^{23} molecules}{mol})\) = 4.99 × 105 molecules ∴ \(\frac{M}{M_0}\) = 4.99. So, t = \(\frac{(4.99 × 10^5)-1}{(1.08 × \frac{10^{-11} cm^3}{s})(1.47 × 10^{14} cm^{-3})(\frac{60s}{min})}\) = 5.24 min and N = \(\frac{N_0 M_0}{M} = \frac{1.47 × 10^{14}}{4.99 × 10^5}\) = 2.95 × 108 \(\frac{particles}{cm^3}\).
10. The structure of protein can be denatured by the organic solvents having high concentration.
a) True
b) False
View Answer
Explanation: The organic solvents have the ability to bind on the protein molecule at a specific position hence leads to the disruption of the interactions caused by hydrophobic bonds and this constitution gives stability to the structure of protein molecule. Therefore, little organic solvent is sufficient for the process of precipitation at low temperature to avoid the denaturation of the protein molecule.
Sanfoundry Global Education & Learning Series – Bioseparation Processes.
To practice all areas of Bioseparation Processes, here is complete set of 1000+ Multiple Choice Questions and Answers.
If you find a mistake in question / option / answer, kindly take a screenshot and email to [email protected]
