This set of Protein Engineering Interview Questions and Answers for freshers focuses on “Post-Translational Modifications – 2”.
1. Which of the following post-translational modification is shown in the figure below?
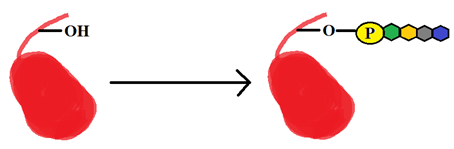
a) S-nitrosylation
b) Ubiquitination
c) Lipidation
d) Phosphoglycosylation
View Answer
Explanation: The post-translational modification shown in the figure above is phosphoglycosylation.
2. Find the odd one out.
a) Glypiation
b) N-glycosylation
c) Phosphoglycosylation
d) S-nitrosylation
View Answer
Explanation: S-nitrosylation is not a type of glycosylation, whereas glypiation, N-glycosylation, and phosphoglycosylation are types of glycosylation. Hence, S-nitrosylation is the odd one out. S-nitrosylation is a critical post-translational modification used by cells to stabilize proteins, regulate gene expression and provide Nitrogen Oxide (NO) donors, and the generation, localization, activation, and catabolism of S-nitrothiols (SNOs) are tightly regulated.
3. Ubiquitin is 8-kDa polypeptide consisting of how many amino acids?
a) 77 amino acids
b) 67 amino acids
c) 66 amino acids
d) 76 amino acids
View Answer
Explanation: Ubiquitin is an 8-kDa polypeptide consisting of 76 amino acids. It is appended to the amino group of lysine in target proteins via its C-terminal glycine. Many such ubiquitin molecules are attached to the target protein to degrade it in the proteasome.
4. Ubiquitination plays a crucial role in the degradation of proteins.
a) False
b) True
View Answer
Explanation: The above statement is true. Ubiquitination plays a crucial role in the degradation of proteins. Following an initial monoubiquitination event, the formation of a ubiquitin polymer may occur. The polyubiquitinated proteins are then recognized by the proteasome complex that catalyzes the degradation of the ubiquitinated protein and the recycling of ubiquitin.
5. S-nitrosylation is an irreversible reaction.
a) True
b) False
View Answer
Explanation: The above statement is false. S-nitrosylation is a reversible reaction, and S-nitrothiols (SNOs) have a short life in the cytoplasm, because of the host of reducing enzymes that denitrosylate proteins.
6. S-adenosyl methionine (SAM) is suggested to be the most used substrate in enzymatic reactions.
a) True
b) False
View Answer
Explanation: The above statement is false. S-adenosyl methionine (SAM) is suggested to be the most used substrate in enzymatic reactions, after ATP. Additionally, while N-methylation is irreversible, O-methylation is potentially reversible.
7. Which of the following post-translational modification aids in converting hydrophobic or lipophilic compounds into hydrophilic compounds?
a) Glycosylation
b) Methylation
c) Ubiquitination
d) Hydroxylation
View Answer
Explanation: Hydroxylation is the post-translational modification that aids in converting hydrophobic or lipophilic compounds into hydrophilic compounds. It is catalyzed by enzymes termed as hydroxylases. This process adds a hydroxyl group (-OH) to the proteins.
8. Which of the following is a well-known mechanism of epigenetic regulation?
a) S-nitrosylation
b) Phosphorylation
c) N-glycosylation
d) Methylation
View Answer
Explanation: Methylation is a well-known mechanism of epigenetic regulation, as histone methylation and demethylation influence the availability of DNA for transcription. Methylation is the addition of methyl groups to amino acid residues in the protein.
9. Which of the following post-translational modification is shown in the figure below?
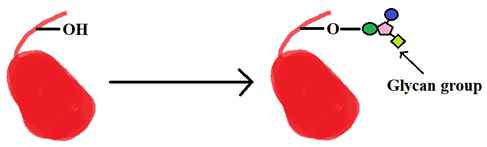
a) Glypiation
b) N-Glycosylation
c) Phosphoglycosylation
d) S-nitrosylation
View Answer
Explanation: The post-translational modification shown in the figure above is S-nitrosylation.
10. Which of the following is a donor of the methyl group in methylation of proteins?
a) Methionine
b) Methane
c) O-adenosyl methionine
d) S-adenosyl methionine
View Answer
Explanation: S-adenosyl methionine is a donor of the methyl group in the methylation of proteins. Methionine, methane, and O-adenosyl methionine are not donors of the methyl group in methylation of proteins. Methylation is mediated by methyltransferases.
11. Which of the following is a method to target proteins to membranes in organelles, vesicles, and plasma membrane?
a) Methylation
b) Glycosylation
c) Phosphorylation
d) Lipidation
View Answer
Explanation: Lipidation is a method to target proteins to membranes in organelles (endoplasmic reticulum, Golgi apparatus, mitochondria), vesicles (endosome, lysosome) and plasma membrane. Methylation, glycosylation, and phosphorylation are not methods to target proteins to membranes in organelles, vesicles, and plasma membrane.
12. Proteolysis is critical to all of the processes listed below except which?
a) Apoptosis
b) Antigen processing
c) Cell signaling
d) Cell growth
View Answer
Explanation: Proteolysis is critical to cellular processes like apoptosis, antigen processing, cell signaling, and surface protein shedding. But it is not critical to cell growth. Proteolysis is a thermodynamically favorable and irreversible reaction. Therefore, proteolysis activity is highly regulated.
13. What is the process in which glutamine is converted to glutamic acid or pyroglutamic acid?
a) Acetylation
b) Methylation
c) Hydroxylation
d) Deamidation
View Answer
Explanation: Deamidation is the process in which glutamine is converted to glutamic acid or pyroglutamic acid. It is the removal or conversion of asparagine or glutamine residue to another functional group. Asparagine is converted to aspartic acid or isoaspartic acid.
14. Which of the following pathway targets lysosomal enzymes to lysosomes?
a) CMP pathway
b) EMP pathway
c) G6P pathway
d) M6P pathway
View Answer
Explanation: Mannose-6-phosphate (M6P) pathway targets lysosomal enzymes to lysosomes. CMP pathway, EMP pathway, and G6P pathway do not target lysosomal enzymes to lysosomes. Multiple N-linked oligosaccharides are added to most lysosomal enzymes in the rough ER and become phosphorylated in Golgi.
15. Which of the following amino acid is most commonly phosphorylated in prokaryotes and plants?
a) Alanine
b) Lysine
c) Glycine
d) Histidine
View Answer
Explanation: Histidine is most commonly phosphorylated in prokaryotes and plants. Alanine, lysine, and glycine are not phosphorylated in prokaryotes and plants. Phosphorylation possibly changes a protein’s structure by altering interactions with nearby amino acids.
Sanfoundry Global Education & Learning Series – Protein Engineering.
To practice all areas of Protein Engineering for Interviews, here is complete set of 1000+ Multiple Choice Questions and Answers.
If you find a mistake in question / option / answer, kindly take a screenshot and email to [email protected]
- Apply for Biotechnology Internship
- Practice Biotechnology MCQs
- Check Biotechnology Books
- Check Protein Engineering Books
