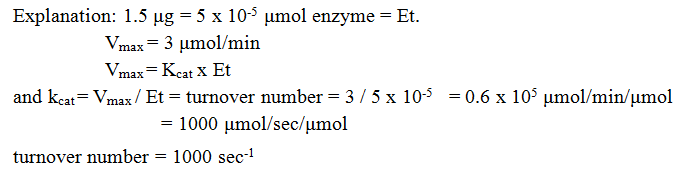This set of Bioprocess Engineering Quiz focuses on “Determining Enzyme Kinetic Constants from Batch Data”.
1. Which stage is preferred for the estimation of the rate of enzymatic reaction?
a) Initial
b) Mid
c) Final
d) Stationary
View Answer
Explanation: Typically, only initial rate data are used. This means that several batch experiments are carried out with different initial substrate concentrations; from each set of data the reaction rate is evaluated at time zero. Initial rates and corresponding initial substrate concentrations are used as (v, s) pairs which can then be plotted in various ways for determination of vmax and Km. Initial rate data are preferred for enzyme reactions because experimental conditions such as enzyme and substrate concentrations are known most accurately at the start of the reaction.
2. Which of the following plot has the difficulty of extrapolating vmax?
a) Line-weaver burk plot
b) Eadie-Hofstee plot
c) Michaelis-menten plot
d) Langmuir plot
View Answer
Explanation: This simple procedure involves plotting (v, s) values directly vmax is the rate as s→∞ and Km is the value of s at v = vmax/2. The accuracy of this method is usually poor because of the difficulty of extrapolating to vmax.
3. Which of the following plot is also known as a double reciprocal plot?
a) Line-weaver burk plot
b) Eadie-Hofstee plot
c) Michaelis-menten plot
d) Langmuir plot
View Answer
Explanation: The double-reciprocal equation is obtained by taking the reciprocal of both sides of the Michaelis-Menten equation. The double-reciprocal (also known as the Lineweaver-Burk) plot is created by plotting the inverse initial velocity (1/V0) as a function of the inverse of the substrate concentration (1/[S]). The vmax can be accurately determined and thus KM can also be determined with accuracy because a straight line is formed. The slope of the resulting line is KM/vmax, the y-intercept is 1/vmax, and the x-intercept is -1/KM.
4. Which plot represents the following equation?
\(\frac{s}{v} = \frac{K_m}{v_{max}} + \frac{s}{v_{max}}\)
a) Line-weaver burk plot
b) Eadie-Hofstee plot
c) Michaelis-menten plot
d) Langmuir plot
View Answer
Explanation: A Langmuir plot of s/v versus s should give a straight line with slope l/vmax and intercept Km / vmax. Linearisation of data for the Langmuir plot minimises distortions in experimental error. Accordingly, its use for evaluation of vmax and Km is recommended.
5. Which plot represents the following equation?
\(\frac{1}{v} = \frac{K_m}{v_{max^s}} + \frac{1}{v_{max}}\)
a) Line-weaver burk plot
b) Eadie-Hofstee plot
c) Michaelis-menten plot
d) Langmuir plot
View Answer
Explanation: This method uses a linearisation procedure to give a straight- line plot from which vmax and Km can be determined. So that a plot of 1/v versus 1/s should give a straight line with slope Km/vmax and intercept l/vmax. This double-reciprocal plot is known as the Lineweaver-Burk plot, and is frequently found in the literature on enzyme kinetics. However, the linearisation process used in this method distorts the experimental error in v, so that these errors are amplified at low substrate concentrations.
6. What is the term “Km”?
a) Concentration of the enzyme
b) Concentration of the catalyst
c) Concentration of the product
d) Concentration of the substrate
View Answer
Explanation: For practical purposes, Km is the concentration of substrate which permits the enzyme to achieve half vmax. An enzyme with a high Km has a low affinity for its substrate, and requires a greater concentration of substrate to achieve vmax.
7. What is the unit of vmax?
a) mmol
b) mol/sec
c) mol
d) mol/hr
View Answer
Explanation: vmax “represents the maximum rate achieved by the system, at maximum (saturating) substrate concentrations”, Unit: μmol/min (or mol/s).
8. Is vmax affected by enzyme concentration?
a) True
b) False
View Answer
Explanation: The reaction rate still increases with increasing substrate concentration, but levels off at a much lower rate. By increasing the enzyme concentration, the maximum reaction rate greatly increases. However, enzymes become saturated when the substrate concentration is high.
9. Km is directly proportional to the rate of enzyme binding?
a) True
b) False
View Answer
Explanation: Km reflects the enzyme’s dissociation constant. A high Km means weak binding (the enzyme likes to dissociate from its substrate), and a low Km means strong binding (it doesn’t like to dissociate from its substrate, meaning that it has a strong affinity for the substrate).
10. What is the unit of catalytic efficiency kcat/Km?
a) conc-1 time-1
b) conc time
c) conc-2time-2
d) conc time-1
View Answer
Explanation: When calculating Kcat, the concentration units cancel out, so Kcat is expressed in units of inverse time. It is the turnover number – the number of substrate molecule each enzyme site converts to product per unit time. The units of Km are those of concentration i.e. mM, mM or Km is the concentration of substrate at which half maximal velocity is observed.
11. Which of the following state represents the condition – “Initial mixing of E+S, while [ES] builds up”.
a) Enzyme is saturated with substrate
b) Pre- Steady state
c) Steady – state
d) Steady-state kinetics
View Answer
Explanation: In the first moment after an enzyme is mixed with substrate, no product has been formed and no intermediates exist. The study of the next few milliseconds of the reaction is called pre-steady-state kinetics.
12. What competes with substrate for active site?
a) Competitive inhibition
b) Uncompetitive inhibition
c) Non-competitive inhibition
d) Pure Non-competitive inhibition
View Answer
Explanation: When a fake substrate binds to the active site of an enzyme, it can’t be processed in the same way and it won’t turn into a product. A fake substrate is called a competitive inhibitor. Competitive inhibitors bind the active site of an enzyme, preventing a real substrate from binding and a product from being formed.
13. The concept of “induced fit” refers to the fact that _______
a) When a substrate binds to an enzyme, the enzyme induces a loss of water (desolvation) from the substrate
b) Substrate binding may induce a conformational change in the enzyme, which then brings catalytic groups into proper orientation
c) Enzyme-substrate binding induces an increase in the reaction entropy, thereby catalyzing the reaction
d) Enzyme specificity is induced by enzyme-substrate binding
View Answer
Explanation: Induced-fit model A proposed mechanism of interaction between an enzyme and a substrate. It postulates that exposure of an enzyme to a substrate causes the active site of the enzyme to change shape in order to allow the enzyme and substrate to bind.
14. A given substrate may be acted upon by a number of different enzymes, each of which uses the same substrate (s) and produces the same product (s). The individual members of a set of enzymes sharing such characteristics are known as-
a) Group specific enzymes
b) Isoenzymes
c) Allosteric enzymes
d) Substrate specific enzymes
View Answer
Explanation: Isozymes (also known as isoenzymes or more generally as multiple forms of enzymes) are enzymes that differ in amino acid sequence but catalyze the same chemical reaction. These enzymes usually display different kinetic parameters (e.g. different KM values), or different regulatory properties. Allozymes represent enzymes from different alleles of the same gene, and isozymes represent enzymes from different genes that process or catalyse the same reaction, the two words are usually used interchangeably.
15. 1.5 μg of enzyme gives a vmax of 3 μmol product produced per minute. What is the turnover number for this enzyme?
a) 100 sec-1
b) 200 sec-1
c) 1000 sec-1
d) 2000 sec-1
View Answer
Sanfoundry Global Education & Learning Series – Bioprocess Engineering.
To practice all areas of Bioprocess Engineering for Quizzes, here is complete set of 1000+ Multiple Choice Questions and Answers.
If you find a mistake in question / option / answer, kindly take a screenshot and email to [email protected]
- Check Bioprocess Engineering Books
- Check Biotechnology Books
- Apply for Biotechnology Internship
- Practice Biotechnology MCQs